Why should you convert your model to NeuroML 2?#
Note: for the latest info, see the NeuroML docs.
The NeuroML documentation has been thoroughly updated recently and should be used as the main guide to why and how to convert your model to NeuroML 2.
Converting models to the simulator independent format NeuroML 2 enables conversion of the model to many different formats, as well as enables many of the advanced features of OSB to be used with your model (e.g. 3D visualisation of cells & networks and extracting information on channel kinetics).
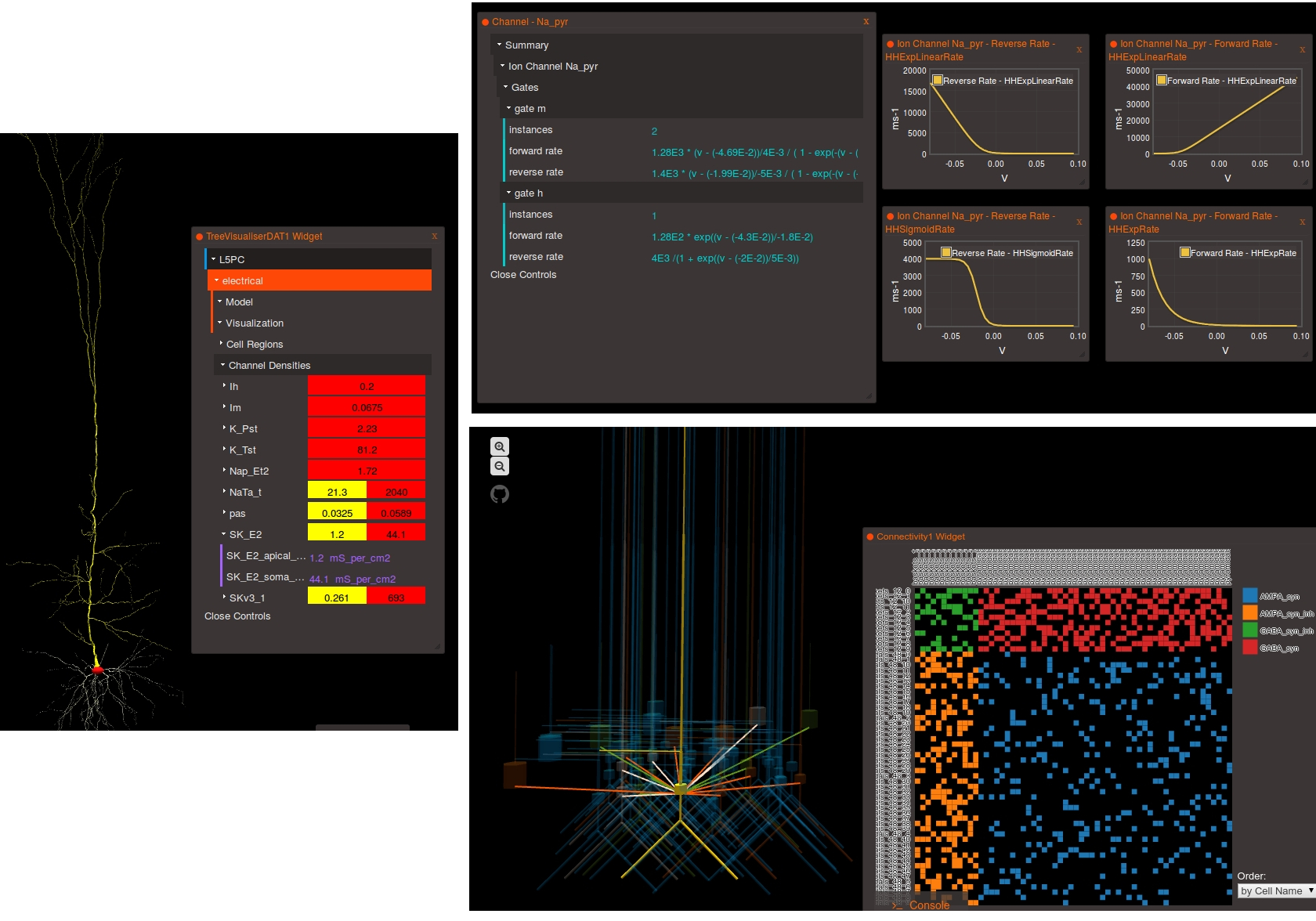
At the moment, there is no “magic button” to press in simulators to export your model into valid NeuroML 2. Presently the best approach is to look at some of the core examples of NeuroML 2, the OSB projects being converted to NeuroML 2, look at the XML examples there and start manual conversion of the models based on these.
There is a step by step guide: Converting conductance based cell models to NeuroML2 here!
There are some suggestions below for tools to use in this process.
Use jNeuroML to validate/execute/convert/view the NeuroML 2 files
jNeuroML can be used to validate NeuroML2 files:
jnml -validate MyNeuroML2.nml
This will validate the file against the latest XSD Schema document, as well as perform a number of other checks on the structure of the model.
Creating a LEMS simulation file to specify how long to run the simulation, timestep, what to plot/save etc. allows the model to be run with jNeuroML. See examples here and most of these OSB projects have at least one LEMS simulation file too.
To quickly visualise cells and networks in NeuroML 2 (as opposed to loading them in neuroConstruct or viewing on OSB), models can be converted to SVG format (for viewing in a web browser etc.):
jnml MyNeuron.cell.nml -svg
jnml MyCircuit.net.nml -svg
Installing the latest development version from source as outlined here is highly recommended to get the latest features of jNeuroML.
Use pyNeuroML to export NeuroML2 morphologies from NEURON
pyNeuroML is a Python package which makes many of the features of jNeuroML accessible in Python scripts by bundling the jNeuroML jar file and using the functionality of that package.
In addition to jNeuroML features, pyNeuroML adds a number of Python specific features, including an updated exporter from NEURON into NeuroML2. This is currently limited to exporting just neuronal morphologies (see example here) but will be developed further to allow extraction of other features such as connectivity (and generation of templates in NeuroML2 for channels, synapses etc.).
This feature was used to extract a number of cell models from the original NEURON code in a 3D olfactory bulb model (see this script).
Use pyNeuroML to help convert channels to NeuroML 2
Another feature being worked on is a set of scripts to help conversion of ion channel models (e.g. in NEURON’s NMODL format) to NeuroML 2. These scripts can be accessed after pyNeuroML is installed using:
pynml-channelanalysis NaConductance.channel.nml # Analyse a NeuroML 2 channel
pynml-modchananalysis NaConductance # Analyse a NEURON channel e.g. from NaConductance.mod
These scripts, while they take different approaches to generating the traces for activation/inactivation variables, produce comparable plots:
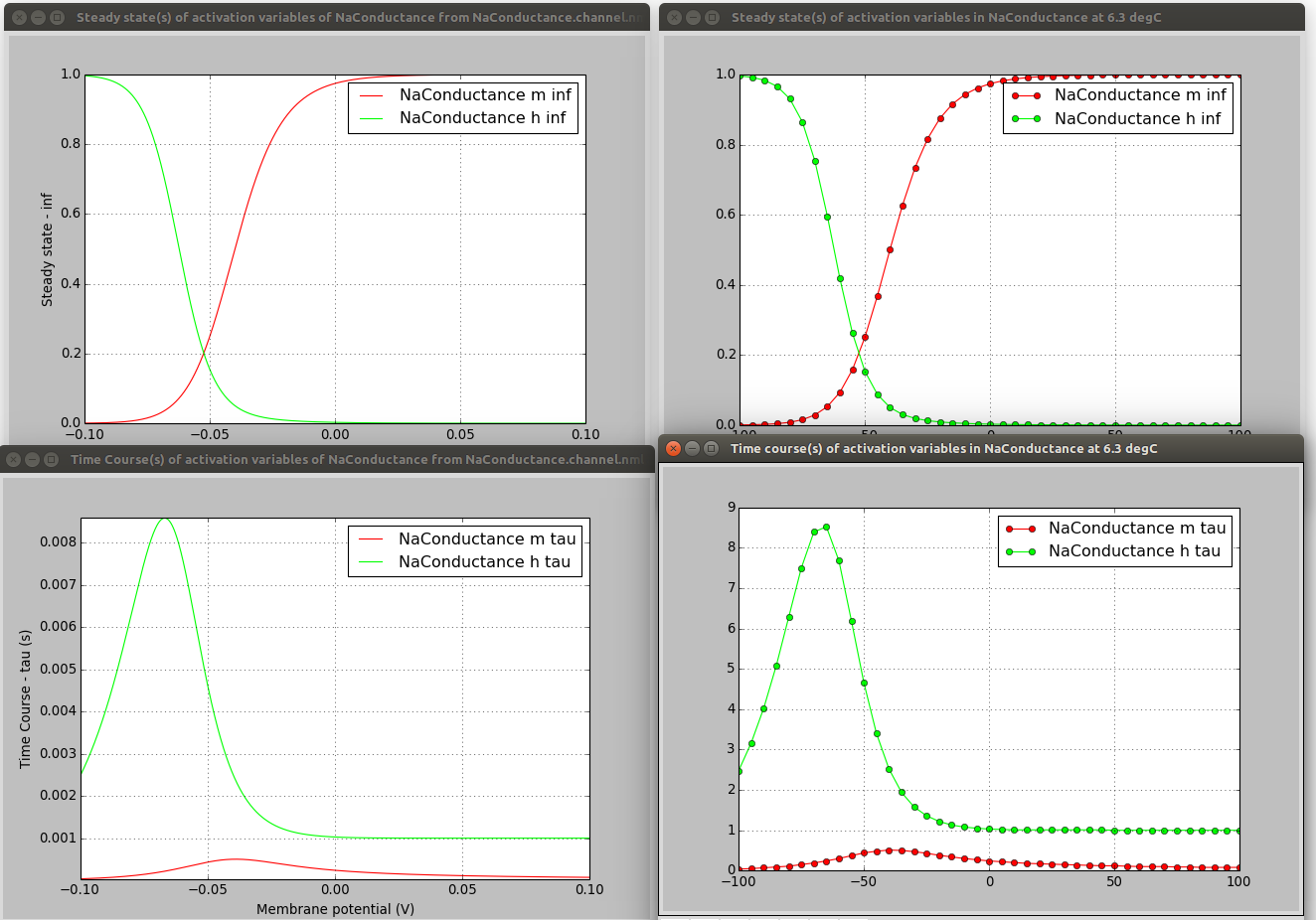
On the left are the plots produced from the NeuroML file (e.g. see here) and on the right from the NMODL version (see here).
To be clear, these scripts don’t (currently) help you extract the equations from the NMODL file and put them in the correct place in the NeuroML file, they just help comparing the behaviour of the 2 channel formats to check for errors in a manual conversion.
Use neuroConstruct to help conversion to NeuroML v1/2
Many projects on OSB were originally converted from the original format (NEURON, GENESIS, etc.) to NeuroML v1.8.1 using neuroConstruct (see here for a list of these), and from there the conversion to NeuroML2 inside neuroConstruct was straightforward. More on using neuroConstruct here and details on conversion of models to NeuroML v1 here.
