Open Source Brain v2#
OSBv2 is live!
The latest release of OSBv2 is available at https://v2.opensourcebrain.org. Please do get in touch if you have any queries or would like to help with user testing.
Open Source Brain (OSB) v2 is a new integrated research platform that builds on the features of OSBv1. It is currently under active development, but is stable enough for users to start using it in their research.
OSBv2 is structured around 3 key concepts: Repositories, Workspaces and Applications.
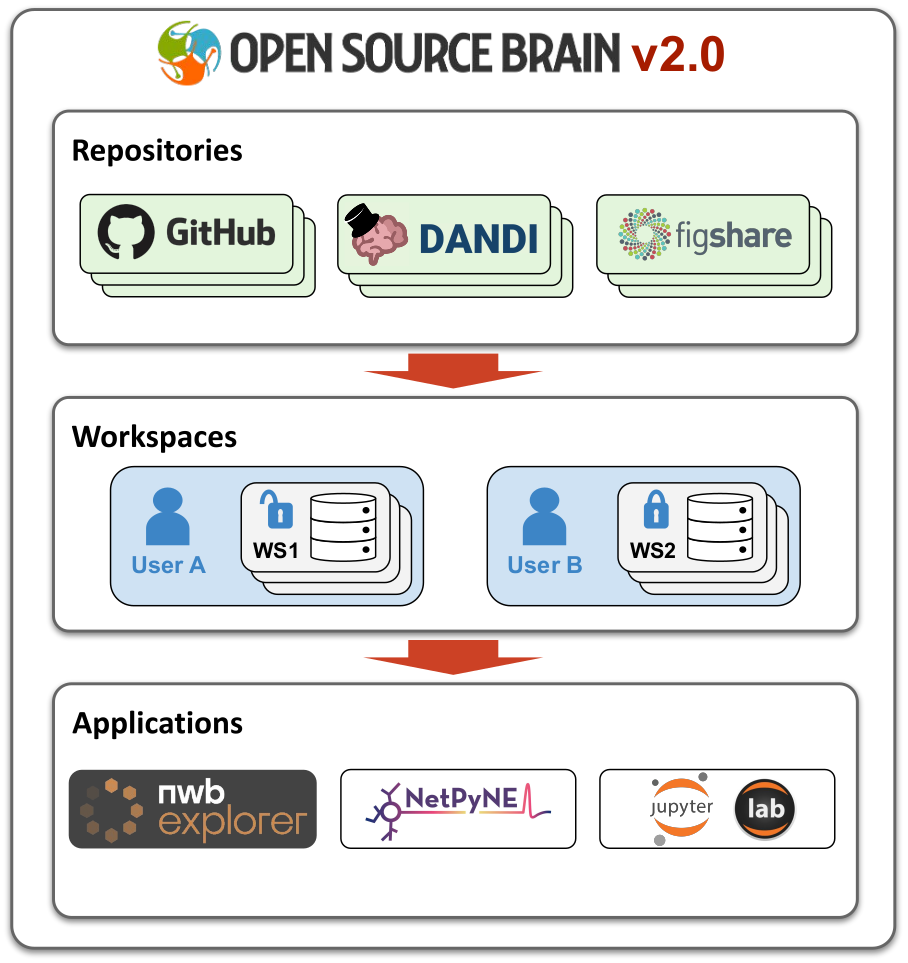
Fig. 1 Relationship between Repositories, Workspaces and Applications on OSBv2.#
Repositories#
Repositories are views of sets of files and data in public resources like GitHub, Figshare and DANDI Archive.
A repository page on OSBv2 lists the current contents of the remote resource (e.g. GitHub repository, FigShare project)
OSB Repositories are public: all users can browse repositories that have been added to OSBv2
Private repositories, and interfaces to other neuroinformatics resources are also planned
More details on repositories can be found here.
Workspaces#
Workspaces are persistent spaces for users to carry out their work in.
Empty workspaces can be created, or they can be seeded from OSBv2 repositories using some or all of the files in the repository at that point in time
Each workspace is saved on a persistent cloud volume, allowing users to save their work and resume it at a later time
Workspaces can be opened in any OSBv2 application, and any files which are generated (e.g. analysis outputs, simulation results) are also stored in the workspace, and other files can be uploaded
Users can keep their workspaces private or make them public to share them with the community
More details on workspaces can be found here.
Applications#
OSBv2 integrates a number of applications for use by researchers, and workspaces can be opened in any of the supported OSBv2 applications:
NWB Explorer: for the interactive exploration/visualisation of the contents of Neurodata Without Borders files
NetPyNE: a graphical user interface for the construction and simulation of neuroscience models
JupyterLab: a complete development environment for the Python programming language
Both NWB Explorer and NetPyNE also include inbuilt Python consoles for ad-hoc scripting and analysis, and a number of Python libraries are pre-loaded into the JupyterLab application for convenience.
More details on OSBv2 applications can be found here.
